Code Optimization: Speed up your code by rearranging data access

Speed is of the essence when it comes to scientific computing. But how do you get your numerical program to run faster? Well, there are many different ways (putting parallel processing or multithreading aside). Besides optimizing the algorithm itself, one of the most effective way is by considering memory access. When a code runs, its data is stored in the computer’s RAM. However, the CPU cannot directly access the RAM. In order to perform a computation, the data first needs to be copied to the cache, a small local memory the CPU has access to. Since the process of copying data between RAM and the cache takes time, the copying operation is designed to grab a larger block surrounding the data of interest. The hope is that the successive calculation will utilize data near-enough to the original piece, and as such, the new data will already reside in the cache. If not, a cache miss results, and another RAM to cache copying operation is performed.
Note, particle codes often have two distinct set of memory optimization requirements: one for the field variables, and one for the particles. Here we consider the fields. The latter one is the topic of a follow up post on efficient particle data structures.
The importance of consecutive memory blocks
In practical terms, this means that when writing code, we need to make sure data components are as much next to each other in memory as possible. This will reduce the number of required RAM to cache transfers. We can illustrate this using a simple example. Let’s say we have a large block of numbers and we want to add them all together. The code will look something like this:
double array[] = new double[size]; for (i=0;i<size;i++) array[i]=i; double sum=0; for (i=0;i<size;i++) sum+=array[i];
This code will give us the optimized performance: the data is located in a single consecutive memory block array. But this isn’t the only way to organize the data. Instead of using a single block, the data could be scattered throughout the RAM and referenced via a linked list. Java makes it easy to utilize linked lists,
LinkedList<Double> linked_list = new LinkedList<Double>(); for (i=0;i<size;i++) linked_list.add(new Double(i)); double sum=0; for (i=0;i<size;i++) sum+=linked_list.get(i);
Now, in this case we had to introduce a bit of additional overhead since we are accessing the data via the get getter, and the data is stored as the Double object instead of double primitive. To make a more faithful comparison, we can rewrite the first array example using the ArrayList,
ArrayList<Double> array_list = new ArrayList<Double>(size); for (i=0;i<size;i++) array_list.add(new Double(i)); double sum=0; for (i=0;i<size;i++) sum+=array_list.get(i);
Although the three codes look quite similar, their performance is anything but! To measure speed, I recorded the start and end time for the summation loop. The actual source code is listed below. In addition, to improve accuracy, the time was measured over 500 iterations of the loop. For size=50*50*50=125,000, the times on my 1.7GHz Intel i7 laptop are:
Array took 0.097051789 seconds ArrayList took 0.411013258 seconds LinkedList took 5692.67988 seconds
In other words, the linked list, which accesses data scattered through out the RAM and as such results in a cache miss on each access, ran 58,000x slower than the consecutive array! Instead of completing in a fraction of a second, the linked list case took almost 95 minutes! (I must admit I did not actually have the patience to wait this long, I extrapolated this time by running only 20 iterations and multiplying the time by 25). This slow down can hardly be attributed to the additional overhead of not using data primitives: the code with the ArrayList took only 3/10ths of a second longer over the code with the primitives.
Real-world example: 3D Finite Difference Laplace Solver
The above example is a bit convoluted, of course. Using a linked list for a simple array like this would be a really silly idea. Let’s now consider a more realistic example, a finite difference solver. Here we consider an extremely simple problem given by the Laplace equation \(\nabla^2 x=0\) and Dirichlet boundaries. Equations of this kind arise commonly in fluid dynamics, diffusion, and electrostatic problems. The discretization of this equation using the finite difference approach and assuming constant mesh spacing \(dx=dy=dz=dh\) is
$$x_{i-1,j,k}+x_{i+1,j,k}+x_{i,j-1,k}+x_{i,j+1,k}+x_{i,j,k-1}+x_{i,j,k+1} – 6x_{i,j,k} = 0
$$
Then using the Gauss-Seidel method, the solver pseudocode becomes:
<for number of iterations / until convergence>
<for all non-boundary nodes>
x[i][j][k] = (1/6.0)*(x[i-1][j][k] + x[i+1][j][k] +
x[i][j-1][k] + x[i][j+1][k] +
x[i][j][k-1] + x[i][j][k+1])
This algorithm gives a solution such as the one shown in Figure 1. For this plot, \(x=100\) was set on the \(i=0\) face and \(x=0\) everywhere else.
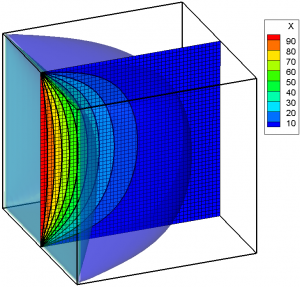
The question is, what is the best way to iterate through the nodes? There are basically two options: i->j->k, or k->j->i. There is no universally correct ordering – the correct choice depends on the way the data is stored in RAM. In this example, the data allocation is such that x(i,j,k)=x[i][j][k],
double x3[][][] = new double[nx][][]; for (int i=0;i<nx;i++) { x3[i] = new double[ny][]; for (int j=0;j<ny;j++) x3[i][j] = new double[nz]; }
First, let’s consider the k->j->i ordering. The solver is then written as:
initData3D(x3); for (it=0;it<num_it;it++) { for (k=1;k<nz-1;k++) for (j=1;j<ny-1;j++) for (i=1;i<nx-1;i++) { x3[i][j][k] = (1/6.0)*(x3[i-1][j][k]+x3[i+1][j][k]+ x3[i][j-1][k]+x3[i][j+1][k]+ x3[i][j][k-1]+x3[i][j][k+1]); } }
For a 100x100x100 array size, this case takes 12.69 seconds to complete on my computer. Next let’s try a simple change: let’s try switching the ordering of the three loops to i->j->k. This gives the following code:
initData3D(x3); for (it=0;it<num_it;it++) { for (i=1;i<nx-1;i++) for (j=1;j<ny-1;j++) for (k=1;k<nz-1;k++) { x3[i][j][k] = (1/6.0)*(x3[i-1][j][k]+x3[i+1][j][k]+ x3[i][j-1][k]+x3[i][j+1][k]+ x3[i][j][k-1]+x3[i][j][k+1]); } }
The only difference between the two codes is the loop ordering. Yet this second case takes a substantially less time to complete: only 3.42 seconds. That’s over 3x speed up from the initial implementation! The speed up becomes even more pronounced as the number of nodes is increased. For a 150x150x150 mesh, the times are 75.73 seconds and 11.68 seconds, a 6.48 times difference. For a 200x200x200 mesh, the speed up reaches 7 times. If this algorithm were typical of the entire simulation program, this speed could result in a code previously taking a week to complete in just one day. The speedup is due to the optimized memory access. The smallest block is the [k] block, which is declared as a single consecutive array. As such, we are better off completing this block before moving to another [i] or [j] location.
Flat vs. 3D array
Three dimensional data can also be stored as a single one-dimensional array. Such a flat array approach becomes handy when matrix operations need to be considered. For instance, the above Laplace problem could also be written as \(\mathbf{L}\vec{x} = 0\), where \(\mathbf{L}\) is the coefficient matrix, containing the discretization of the Laplacian. The size of the flat array is nx*ny*nz. The three dimensional index can be mapped to the flat index using a scheme such us \(u=i*ny*nz + j*nz + k\). The above example, with the faster i->j->k ordering, can then be written as
double x1[] = new double[nx*ny*nz]; initData1D(x1); /*precompute node offsets*/ int node_offset[] = nodeOffsets(); start = System.nanoTime(); for (it=0;it<num_it;it++) { for (i=1;i<nx-1;i++) for (j=1;j<ny-1;j++) for (k=1;k<nz-1;k++) { int u=IJKtoU(i,j,k); sum=0; for (int t=0;t<6;t++) sum+=x1[u+node_offset[t]]; x1[u]=(1/6.0)*sum; } }
In this example, an additional optimization was performed by pre-computing the offsets to the neighbor nodes. These offsets are used to refer to the nodes comprising the standard finite difference stencil. With u=[i][j][k], data at [i-1][j][k] is located at u-ny*nz, using the indexing scheme from above. Similarly, data at [i][j+1][k] is located at u+nz. These node offsets are function of mesh topology and do not change with u. They can thus be precomputed. This will save 6*nx*ny*nz calculations per iteration, which for large meshes can be significant. The computational times for this 1D flat case are listed below, along with the other 2 cases and speed ratios. Although this 1D flat case runs slower than the 3D array, the slow down is nowhere near as dramatic as what was seen with the reversed loop ordering.
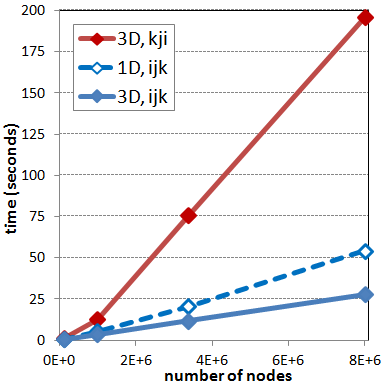
nn | nodes | 3D,ijk | 3D,kji | 1D,ijk | r1 | r3 ------------------------------------------------------ 50 | 125000 | 0.42 | 1.08 | 0.61 | 2.57 | 1.45 100 | 1000000 | 3.42 | 12.69 | 5.41 | 3.71 | 1.58 150 | 3375000 | 11.68 | 75.73 | 20.51 | 6.48 | 1.75 200 | 8000000 | 27.74 | 195.91 | 54.14 | 7.06 | 1.95
These timing studies are also shown graphically in Figure 2 below. Here the two blue curves correspond to the cases with the i->j->k ordering while the red curve is for k->j->i. The dashed blue line shows the flattened 1D array. Comparing the difference between the blue and the red line shows just how important selecting the correct loop ordering really is!
Summary
The way data is organized in memory can have a huge impact on code performance. In this article several alternative methods for storing and accessing data were considered. Using a linked list instead of a consecutive data array resulted in the same algorithm taking 58,000x longer! Even for a more realistic example concerning a finite difference solver, simply rearranging the loops via which the data is accessed resulted in a 7x speed up for large meshes. Yet, the real lesson here is simply the importance of considering performance during code design. Real simulation codes are orders of magnitude more complex than the simple test cases shown here. In such codes, often several different algorithms compete for memory access, and rearranging one can have unintended consequences on the performance of the others. In large codes, your best bet is to use profiling, a timing tool found in most modern development environments (such as Visual Studio, Eclipse, or Netbeans). Profiling will allow you to determine which functions are taking the longest time. In addition, timing studies obtained from the profiler or from a direct instrumentation come in handy in determining just how effective, if at all, an algorithm rewrite was. From my own experience, sometimes rewrites which I thought would definitely improve performance, ended up doing exactly the opposite.
Complete Source Code Listing
package speed;
import java.io.FileWriter;
import java.io.PrintWriter;
import java.util.ArrayList;
import java.util.LinkedList;
/* ************************************************************* Code to test the performance of a finite difference* Poisson solver based on memory access ordering** For more information see:* https://www.particleincell.com/2012/memory-code-optimization/** ***********************************************************/public class Speed
{final static int nn = 50;
final static int num_it = 500;
final static int nx=nn,ny=nn,nz=nn;
public static void main(String[] args)
{int i,j,k;
int it;
long start, end;
/*array*/int size = nx*ny*nz;
double array[] = new double[size];
for (i=0;i<size;i++)
array[i]=i;
double sum=0;
start = System.nanoTime();
for (it=0;it<num_it;it++)
for (i=0;i<size;i++)
sum+=array[i];
end = System.nanoTime();
System.out.println("Array took "+(1e-9)*(end-start)+" seconds");
/*array list*/ArrayList<Double> array_list = new ArrayList<Double>(size);
for (i=0;i<size;i++)
array_list.add(new Double(i));
sum=0;
start = System.nanoTime();
for (it=0;it<num_it;it++)
for (i=0;i<size;i++)
sum+=array_list.get(i);
end = System.nanoTime();
System.out.println("ArrayList took "+(1e-9)*(end-start)+" seconds");
/*linked list*/sum = 0;
LinkedList<Double> linked_list = new LinkedList<Double>();
for (i=0;i<size;i++)
linked_list.add(new Double(i));
start = System.nanoTime();
for (it=0;it<20;it++)
{for (i=0;i<size;i++)
sum+=linked_list.get(i);
}end = System.nanoTime();
System.out.println("LinkedList took "+(1e-9)*(num_it/it)*(end-start)+" seconds");
start = System.nanoTime();
for (it=0;it<num_it;it++)
for (i=0;i<size;i++)
array[i]=i;
end = System.nanoTime();
System.out.println("Array took "+(1e-9)*(end-start)+" seconds");
/*data structures for 3D and 1D approaches*/double x3[][][] = allocate3D();
double x1[] = allocate1D();
/*case 1, 3D k->j->i*/initData3D(x3);
start = System.nanoTime();
for (it=0;it<num_it;it++)
{for (k=1;k<nz-1;k++)
for (j=1;j<ny-1;j++)
for (i=1;i<nx-1;i++)
{x3[i][j][k]=(1/6.0)*(x3[i-1][j][k]+x3[i+1][j][k]+
x3[i][j-1][k]+x3[i][j+1][k]+
x3[i][j][k-1]+x3[i][j][k+1]);
}}end = System.nanoTime();
System.out.println("Case 1, 3D k->j->i took "+(1e-9)*(end-start)+" seconds");
output3D("case1.dat",x3);
/*case 2, 3D i->j->k*/initData3D(x3);
start = System.nanoTime();
for (it=0;it<num_it;it++)
{for (i=1;i<nx-1;i++)
for (j=1;j<ny-1;j++)
for (k=1;k<nz-1;k++)
{x3[i][j][k]=(1/6.0)*(x3[i-1][j][k]+x3[i+1][j][k]+
x3[i][j-1][k]+x3[i][j+1][k]+
x3[i][j][k-1]+x3[i][j][k+1]);
}}end = System.nanoTime();
System.out.println("Case 2, 3D i->j->k took "+(1e-9)*(end-start)+" seconds");
output3D("case2.dat",x3);
/*case 3, 1D flat array*/initData1D(x1);
/*precompute node offsets*/int node_offset[] = nodeOffsets();
start = System.nanoTime();
for (it=0;it<num_it;it++)
{for (i=1;i<nx-1;i++)
for (j=1;j<ny-1;j++)
for (k=1;k<nz-1;k++)
{int u=IJKtoU(i,j,k);
sum=0;
for (int t=0;t<6;t++) sum+=x1[u+node_offset[t]];
x1[u]=(1/6.0)*sum;
}}end = System.nanoTime();
System.out.println("Case 3, flat 1D took "+(1e-9)*(end-start)+" seconds");
output1D("case3.dat",x1);
}/**allocates 3D nn*nn*nn array*/static double[][][] allocate3D()
{double x[][][]=new double[nx][][];
for (int i=0;i<nx;i++)
{x[i] = new double[ny][];
for (int j=0;j<ny;j++)
x[i][j] = new double[nz];
}return x;
}/**resets data, assumes uniform 3D nn*nn*nn mesh*/static void initData3D(double x[][][])
{int i,j,k;
/*set everything to zero*/for (i=0;i<nx;i++)
for (j=0;j<ny;j++)
for (k=0;k<nz;k++)
x[i][j][k]=0;
/*set default value of 100 on x=0 plane*/for (j=0;j<ny;j++)
for (k=0;k<nz;k++)
x[0][j][k]=100;
}/**allocates 1D nn*nn*nn array*/static double[] allocate1D()
{/*allocate memory structure*/return new double[nn*nn*nn];
}/**resets data, assumes uniform 3D nn*nn*nn mesh*/static void initData1D(double x[])
{int i,j,k;
/*set everything to zero*/for (i=0;i<nx;i++)
for (j=0;j<ny;j++)
for (k=0;k<nz;k++)
{x[IJKtoU(i,j,k)]=0;
}/*set default value of 100 on x=0 plane*/for (j=0;j<ny;j++)
for (k=0;k<nz;k++)
{x[IJKtoU(0,j,k)]=100;
}}/**flattens i,j,k index, u = i*(ny*nz)+j*(nz)+k*/static int IJKtoU(int i, int j, int k)
{return i*(ny*nz) + j*nz + k;
}/**returns node offsets for a standard finite difference stencil*/static int[] nodeOffsets()
{int node_offsets[] = new int[6];
node_offsets[0]=IJKtoU(-1,0,0);
node_offsets[1]=IJKtoU(+1,0,0);
node_offsets[2]=IJKtoU(0,-1,0);
node_offsets[3]=IJKtoU(0,+1,0);
node_offsets[4]=IJKtoU(0,0,-1);
node_offsets[5]=IJKtoU(0,0,+1);
return node_offsets;
}/**saves 3D mesh in the Tecplot format*/static void output3D(String file_name, double x3[][][])
{PrintWriter pw = null;
try{
pw = new PrintWriter(new FileWriter(file_name));
}catch (Exception e)
{System.err.println("Failed to open output file "+file_name);
}pw.println("VARIABLES = i j k X");
pw.printf("ZONE I=%d J=%d K=%dn",nx,ny,nz);
for (int i=0;i<nx;i++)
for (int j=0;j<ny;j++)
for (int k=0;k<nz;k++)
pw.printf("%d %d %d %gn",i,j,k,x3[i][j][k]);
pw.close();
}/**saves flat 1D mesh in the Tecplot format*/static void output1D(String file_name, double x1[])
{PrintWriter pw = null;
try{
pw = new PrintWriter(new FileWriter(file_name));
}catch (Exception e)
{System.err.println("Failed to open output file "+file_name);
}pw.println("VARIABLES = i j k X");
pw.printf("ZONE I=%d J=%d K=%dn",nx,ny,nz);
for (int i=0;i<nx;i++)
for (int j=0;j<ny;j++)
for (int k=0;k<nz;k++)
pw.printf("%d %d %d %gn",i,j,k,x1[IJKtoU(i,j,k)]);
pw.close();
}}
You can also download the source code. And do not hesitate to contact us if you have an old code that could benefit from some optimization. Here at PIC-C, we have many years of experience analyzing and optimizing simulation codes and will gladly apply our experience towards your problem.
References
- Class notes, Virginia Tech CS4414, “Issues in Scientific Computing”, taught by Adrian Sandu, ~2003
- Wadleigh, K.R., and Crawford, I.L., “Software Optimization for High Performance Computing: Creating Faster Applications”, Prentice Hall, 2000
What is the operation count of Gauss-Seidel for 1D, 2D and 3D matrices of Laplace equation. Using Ordinary Elimination to solve the problem requires order N in 1D and N^4 in 2D and N^7 in 3D ( order * bandwidth^2 = N^2 * (N^2)^2 = N^7 ). The Fast Poisson Solver or FFT requires N^2 * log2(N) operations in 2D and I think N^3 * log2(N^2) in 3D ( order * log2(bandwidth) ). So for a 3D matrix with N=100, using the FFT would would be N^4 / log2(N^2) faster or over 7 Million times faster than ordinary elimination. I wonder how much faster it would be compared to 3D Gauss-Seidel. Here is an MIT lecture on Fast Poisson Solver on a square or cubed shape.
http://ocw.mit.edu/courses/mathematics/18-086-mathematical-methods-for-engineers-ii-spring-2006/video-lectures/lecture-20-fast-poisson-solver/
and the corresponding book chapter
http://www.myoops.org/twocw/mit/NR/rdonlyres/Mathematics/18-086Spring-2005/EC160E25-0F93-4D53-A297-4FDDD4E44AC6/0/am72.pdf
Correction, the FFT operation count is Mlog2(M) . So for 1D let M = N, for 2D M = N*N and for 3D M = N*N*N.
In 2D the FFT operation count is
N^2 * log2(N^2) = 2 * N^2 * log2(N)
In 3D the FFT operation count is
N^3 * log2(N^3) = 3 * N^3 * log2(N)
For a 3D mesh with N = 64 the FFT speedup over Ordinary Elimination is N^7 / (3*N^3*log2(N)) = 932067 times faster.
For a 3D mesh with N = 128 the FFT is 12782640 times faster than ordinary elimination algorithm.
I wonder how much faster the FFT or Fast Poisson Solver will be than using Gauss-Seidel in 3D. It would be interesting to implement it to compare and see what the effect of cache misses has on it.
John, from my own experience, such highly optimized matrix solvers are not all that usable in practice. Especially when you consider algorithms such as the particle in cell. In PIC you end up doing bunch of other things besides solving matrixes, such as pushing particles. Often, the time spent pushing particles exceeds the matrix solver by an order of magnitude. So even if you get a super optimized solver, you are still buying just so much. My take on code optimization is to understand the basics (such as memory access) and apply these lessons across the board. I think that in general this works better than focusing all your energy on a single component as folks who write these super optimized solvers do.
But I agree, Gauss Seidel is not a very good algorithm. In my codes I generally use a diagonally-preconditioned conjugate gradient. I didn’t use it in this article since the algorithm is more complicated. A PCG solver can often converge in sqrt(n) steps which is a huge improvement over the n*log(n) steps (or so) needed by GS. The beauty of GS is that it’s very simple to implement and can thus also be used to verify more complicated solvers such as the PCG.
I looked it up and iterative solvers like GS have an operation count of
count = s * N * M
where
M is the number of iterations required to converge.
N is the number of unknowns.
s is the number of nonzero coefficients per equation.
Using the above symbols the FFT operation count is N * log2(N). So the speedup of using FFT over GS would be s*M / log2(N). Taking s to be 1 and M = 500, N = 100^3, then FFT is 25 times faster than GS.
When you have a rectangle shape in 2D or box shape in 3D then I think the FFT method is supposed to be the fastest solver.
I found the operation count for iterative solvers in table 3.1 in the link below.
http://books.google.ca/books?id=oDo3LqUa6bgC&pg=PA30&lpg=PA30&dq=Gauss+Seidel+operation+count&source=bl&ots=6S1v36x-Qf&sig=vBZoHG6u1ZXIlXmW-iXHOxKCLmI&hl=en&sa=X&ei=b3-XT4LBMrHMiQLi-q0S&ved=0CFIQ6AEwBg#v=onepage&q=Gauss%20Seidel%20operation%20count&f=false
This shows you just how important selecting the right algorithm is. The algorithm should be always the first thing to consider when optimizing your code. There are generally always multiple ways of getting something done, and some will be inherently faster than others. Only after you pick the fast algorithm you should start playing with rewriting memory access, unrolling loops, and so on…
By the way, that link didn’t work for me. It says the page is not available. I think Google Books picks randomly which pages show up and I guess this wasn’t one of them.
The link works fine for me each time. Anyway it is section 3.4 Operation Counts in the book if you want to look up the book in your library. In it they provide a couple of tables of operation counts in 1D, 2D and 3D for inversion of a matrix, LU decomposition and iterative methods. They also say that the value of “s” in the formula I gave is 3 for 1D , 5 for 2D and 7 for 3D. This matches your equation for 3D because you are doing 6 adds and 1 multiply for each unknown in the equation above your figure 1 in this blog.
They go on to say this about 3D.
“Many important applications today are fundamentally 3-D, and memory requirements alone are making iterative methods necessary. There is a big window of oportunity in the 3-D iteration count, from n^7 (the operation count for 3-D LU) to Mn^3. Finding practical iterative methods which have M ~ n^4 is a major frontier in 3-D. Recall that N = n^3 for 3D; so a practical 3D target is achieving M ~ N^(4/3) .”
I have a question – Typically in a PIC code how much efficiency (speed up) can be achieved by using single precision instead of double precision and how much will I compromise on accuracy. Is there some typical estimation on this….or some paper.
You know, I am not sure how much of this still holds. There are basically two reasons why single precision would give you a faster code. First, if I am not mistaken, older CPUs were optimized to deal with single precision numbers, and as such operations on doubles were inherently slower. I think this is no longer the case. However, the second speed up still exists and is related to the point of this article. If you use singles, you will end up using half the memory space, and as such, you’ll be able to fit more data in the cache, resulting in faster memory access. And of course you can also run larger cases.
We generally use doubles now. I’ll look into the effects in a future article, it’s definitely interesting. Thanks for the suggestion!
The problem with single precision is that you can resolve only about 7 orders of magnitude difference. Although this may seem like a lot, it is quite easy to get to this threshold when for instance scattering particle charges to the grid, especially if you have variable particle weights. Just after scattering few massive particles, scattering a large number of small particles may not increase the charge density any further. The solution would then be to scatter the light particles first, however, that is often not practical.
Thanks Ludos for the reply, I agree with your comments……..I am looking forward to your answer for a typical test case just to have a quantitative idea about the gain in speed.
I performed this test for my PIC code for both the cases – with single precision and double precision. For the particular problem I tested single precision code took 80% of the time taken by a double precision code. So typically we can say single precision codes should be around 20-25% faster. However definitely there is an accuracy issue, so it is always better to use a double precision version.
Thanks for the info!